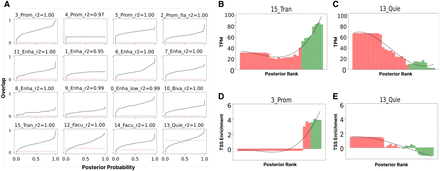
Posterior probability is associated with reproducibility. (A) The frequency of overlap with a corresponding state as a function of posterior probability for all states in the base annotation in the running example (ChromHMM, GM12878, S1). In each subpanel of A, the horizontal axis corresponds to the posterior probability, and the vertical axis corresponds to the observed overlap. The red dotted horizontal line in each subpanel represents the genome coverage of the corresponding state in the verification state, indicating the overlap expected from random overlap. The R2 score represents how well the isotonic regression curve fits the data. (B) The mean expression level (TPM) as a function of the posterior of 15_Tran(B). The horizontal axis represents rank of posterior probability, and the vertical axis shows TPM. (C) Same as B, but for 13_Quie(B). Similarly, D shows the enrichment around TSS regions as a function of the posterior of a 3_Prom(B). The horizontal axis shows the rank of posterior probability, and the vertical axis shows enrichment around TSS. (E) Same as D, but for 13_Quie(B); confirming that the posterior contains biologically relevant information about transcription start sites. In B–E, green bars represent genomic loci for which the maximum-a-posterior is the target chromatin state in the base annotation, and red bars correspond to the loci where the maximum-a-posterior is another chromatin state.











