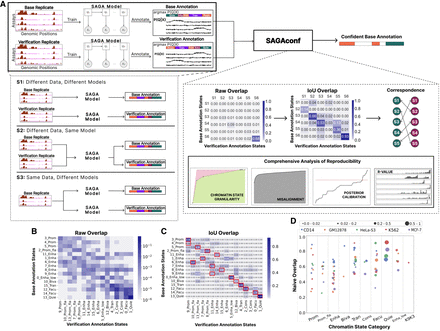
Schematic workflow. (A) We obtained histone modification assays from biological replicates via ENCODE DCC (The ENCODE Project Consortium 2012) and used these data sets to train SAGA models (Segway or ChromHMM) to generate chromatin state annotations. The SAGA model outputs a matrix representing the posterior probability P(Q|X) values of assigning each chromatin state to each genomic position and a vector of state labels assigned to the position with the highest posterior probability, argmaxP(Q|X) (Methods). One set of replicated data is chosen as the base and the other as the verification. SAGA training and genome annotation are performed according to three settings of variability: S1 (different data, different models), in which two separate SAGA models are trained independently using data from each biological replicate; S2 (different data, same model), in which data from both replicates are concatenated to train a single SAGA model that provides separate annotations for each replicate; and S3 (same data, different models), in which the same data set (base replicate) is used to train two different SAGA models with different parameter initializations. Both the base and verification annotations, generated by any variability setting, are inputs to SAGAconf. The SAGAconf evaluation pipeline begins by forming a pairwise overlap frequency distribution matrix between the two annotations and calculating the intersection over union (IoU) overlap to determine the correspondence between state pairs across the annotations. SAGAconf performs reproducibility analysis and outputs a subset of the base annotation that it identifies as confident (Methods). (B) The raw overlap frequency distribution from our running example annotation (S1; ChromHMM, GM12878). Rows and columns correspond to states in base and verification annotations, respectively. Color indicates frequency of overlap (log scale). (C) Same as B, but color indicates the IoU of overlap is derived from a raw overlap matrix (linear scale). For each chromatin state of the base annotation, its corresponding state in the verification annotation is defined as the one with the maximum IoU (marked with red square). (D) Fraction of overlap (Naive overlap) of various chromatin states categories identified in the ChromHMM annotations according to S1 for all five cell types. Each dot represents a chromatin state, with color denoting cell type and size proportional to genome coverage.











