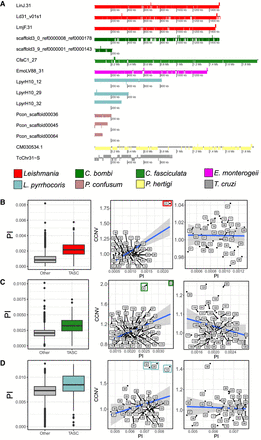
TASC has almost all genes in the same coding strand and an increased nucleotide diversity. (A) Gene distribution of all TASC. Each box corresponds to a gene, and the position of the box below or above the strand corresponds to the gene orientation. The colors represent the species/clades of origin of each chromosome. All the genes are usually in the same coding strand. Nucleotide diversity and comparison between diversity and CCNV for the L. donovani EA_HIV set (B), C. bombi (C), and Leptomonas (D). Left panels are box plots representing 10-kb window π-values comparing TASC and other chromosomes. Chromosomes from TASC are colored, whereas others are gray. (Middle) Correlation between CCNV (y-axis) and π (x-axis) in all chromosomes. (Right) Without TASC. The TASC are highlighted by colored boxes. L. donovani EA_HIV set: (middle) r = 0.719, P-value 7.58 × 10−7; (right) r = −0.001, P-value = 0.9943. C. bombi: (middle) r = 0.429, P-value 0.001082, (right) r = −0.229, P-value = 0.09815. Leptomonas: (middle) r = 0.309, P-value 0.05542; (right) r = −0.133, P-value = 0.4392.











