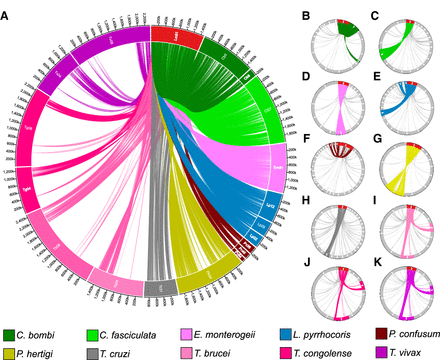
Chromosomes with a high density of orthologous genes with Leishmania Chromosome 31 (Leish Chr 31) are also present extra copies in other trypanosomatids. Circa plots representing the orthology between Leish Chr 31 (red box) and chromosomes from other species (colored boxes), drawn in proportion to their sizes. The presence of ortholog genes between Leish Chr 31 and a given chromosome is shown by linking lines. (A) Gene sharing between Leish Chr 31 and chromosomes that are consistently supernumerary in other clades (TASC) and in Chromosome 4 and 8 from T. brucei and closely related clades. (B–K) All orthologs between Leish Chr 31 genes and genes in any chromosome from other clades, separated by species. Only chromosomes that share at least one ortholog gene with Leish Chr 31 are shown. Chromosomes that are consistently supernumerary are highlighted by colors, whereas other chromosomes are in light gray. (B) Crithidia bombi; (C) Crithidia fasciculata; (D) Endotrypanum; (E) Leptomonas; (F) Paratrypanosoma; (G) Porcisia; (H) Trypanosoma cruzi; (I) Trypanosoma brucei; (J) Trypanosoma congolense; (K) Trypanosoma vivax. Larger versions of these plots can be seen in the Supplemental Figure S4.











