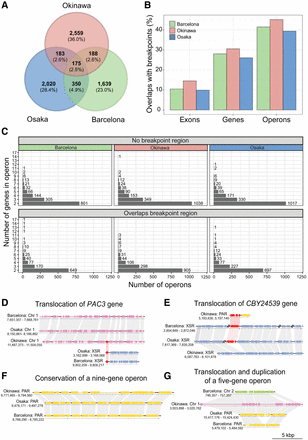
Conservation of operons in O. dioica lineages using the chromosome-level genomes as representatives. (A) Number of shared and unique operons across the chromosome assemblies representing each lineage. (B) The proportion of protein-coding genetic elements that overlap a breakpoint region for each genome. (C) Size distribution of operons that overlap or do not overlap a breakpoint region. (D,E) Translocation of the genes PAC3 (D) and CBY24539 (E; putative activin type I receptor), belonging to different operons in Okinawa lineages and the other lineages. (F) The nine-gene operon reported by Ganot et al. (2004) is conserved in Osaka, Barcelona, and Okinawa. (G) An example of an operon that has been translocated to different chromosomes in each species and duplicated in the Barcelona genome.











