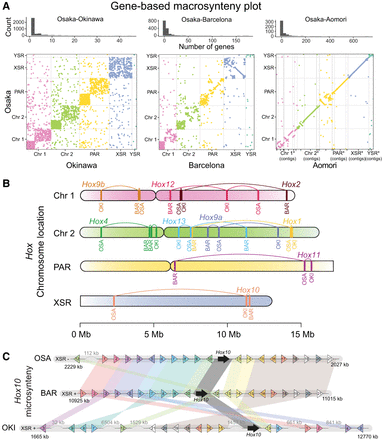
The preservation of orthologous synteny blocks gradually decreases with increasing evolutionary distance in O. dioica. (A, top) Histogram of the number of orthologous genes per syntenic region in pairs of genomes. (Bottom) Dot plots indicating the coordinates of genes belonging to the same orthogroup in pairs of genomes. (B) Comparative chromosome mapping of the Hox genes in the genomes of O. dioica from Osaka (OSA), Barcelona (BAR), and Okinawa (OKI). (C) Comparative microsynteny conservation of the block of the next 10 genes at each side of the Hox10 genes in the same three genomes.











