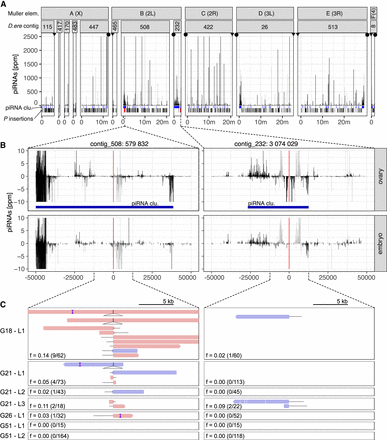
At least two P-element insertions in germline clusters are present in replicate 2 at early stages of the invasion (generations 18–26). (A) Overview of ambiguously (light gray) and unambiguously (black) mapping piRNA in 1-kb windows along the 12 largest contigs of the D. erecta assembly. The corresponding Muller element and the likely direction of the telomere (triangle) and the centromere (circle) are shown. At the bottom, we show the positions of the annotated piRNA clusters (blue) of P-element insertions outside (black) and inside of piRNA clusters (red bold; solely insertions in replicate 2 at generations 26 or less supported by at least two long reads are shown). (B) Abundance of piRNAs in a 100-kb window around the two insertions in piRNA clusters for both ovaries and embryos. Sense piRNAs are on the positive y-axis and antisense piRNAs on the negative y-axis. Ambiguously (light gray) and unambiguously (black) mapped reads are shown. (C) Support of the P-element insertions by long reads at different generations (Gxx) and in different sequencing libraries (Lx). Sense reads are shown in red, and antisense reads are in blue. Regions of the reads aligning to the P-element are shown as dashed lines (true to scale). For each library, we indicate the population frequency of the P-element insertion (f), the number of reads supporting the insertion, and coverage at the insertion site (in parentheses: reads/coverage). Note that both insertions are likely full-length P-element insertions (almost the complete P-element is covered by some reads) and that both insertions are likely lost (or at very low frequency) by generation 51.











