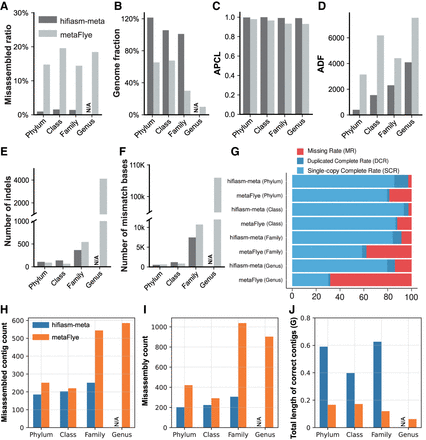
Comparison of hifiasm (dark gray) and metaFlye (light gray) on data set at the phylum, class, family, and genus levels. (A) Misassembled contig rate from MetaQUAST. (B) Genome fraction from MetaQUAST. (C) Average proportion of largest category (APLC). (D) Average distance difference (ADF). (E) Number of indels per 100 kbp from MetaQUAST. (F) Number of mismatches per 100 kbp from MetaQUAST. (G) Unique k-mer-based complete rates (including single complete rate, duplicated complete rate, and missing rate). At the genus level, MetaQUAST was not able to generate evaluation results for the hifiasm-meta, leading to the missing bars in the figures. (H) Misassembled contig count in different taxonomic levels. (I) Misassembly (a concept defined by QUAST representing the breakpoint in the alignment between assembly of reference) count in different taxonomic levels. (J) Total length of correct contigs in different taxonomic levels. At the genus level, owing to the ultra-high computational consumption, QUAST result of hifiasm-meta cannot be obtained within limited time and thus is represented as “N/A.”











