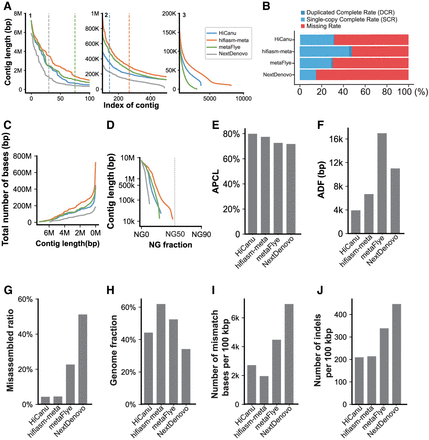
Summary of the performances of HiCanu, hifiasm-meta, metaFlye, and NextDenovo on a synthetic data set. (A) Sorted contig length (left: index 1–100; middle: index 1–500; right: index 1–10,000); the dashed line represents the N50 contig length. (B) Single-copy completeness rate (SCR) and duplicated completeness rate (DCR). (C) Cumulative assembly size as a function of the minimum contig length allowed in the assembly. (D) NGx length (the dashed line denotes the NG50). (E) Average proportion of largest category (APLC). (F) Average distance difference (ADF). (G) Misassembled contig rate computed by MetaQUAST. (H) Genome fraction computed by MetaQUAST. (I) Number of mismatches per 100 kbp computed by MetaQUAST. (J) Number of indels per 100 kbp computed by MetaQUAST.











