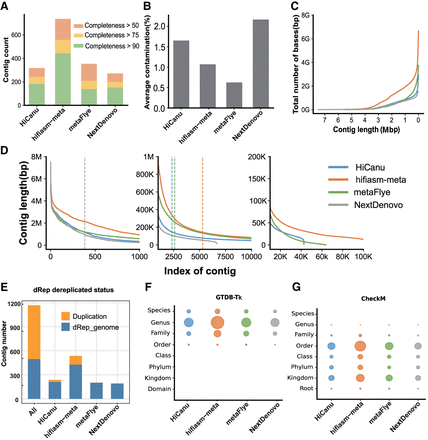
Summary of the performances of HiCanu, hifiasm-meta, metaFlye, and NextDenovo on the sheep gut metagenome data set. (A) Number of assembled contigs (contamination < 10%) for different levels of completeness; completeness was calculated by CheckM. (B) Average contamination rate calculated by CheckM. (C) Cumulative assembly size as a function of the minimum contig length allowed in the assembly. (D) Sorted contig length (left: longest 1000; middle: index 2500–10,000; right: index 20,000–100,000). (E) Duplication analysis. The orange bars indicate the number of duplicate contigs removed by the tool dRep; the blue bars represent the nonredundant contigs. (F) Taxonomic classification of contigs >1 Mbp with GTDB-Tk; the size of each circle represents the number of contigs identified at each taxonomic level. (G) Taxonomic classification of contigs >1 Mbp with CheckM; the size of each circle represents the number of contigs identified at each taxonomic level.











