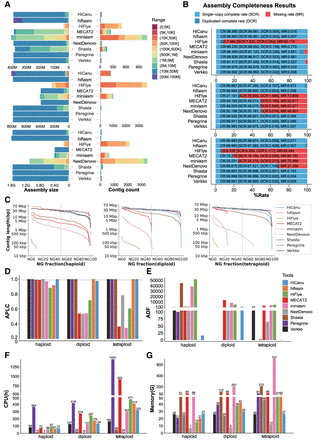
Summary of the performances of the selected genome assemblers on synthetic HiFi reads for rice in haploid form, synthetic diploid, and synthetic tetraploid. (A) Cumulative total size of contigs (left) and the number of contigs (right) that have a size in the range encoded by the color in the legend (top: haploid rice; middle: synthetic diploid rice; bottom: synthetic tetraploid rice); the vertical dashed line indicates the expected genome size. (B) Single complete rate, duplicated complete rate, and missing rate (top: haploid rice; middle: synthetic diploid rice; bottom: synthetic tetraploid rice). (C) Contig length distribution for various choices of the NGx fraction threshold. (D) Average proportion of largest category (APLC); the horizontal dashed line is APLC = 0.99. (E) Average distance difference (ADF). (F) Running time analysis. (G) Memory usage analysis (legend on panel E also applies to panels F,G).











