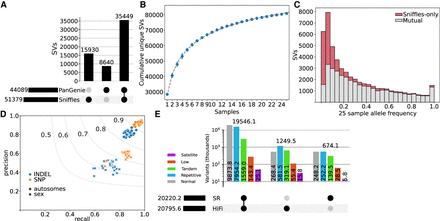
Comparison of variant calling with a small long-read cohort. (A) SV intersection between PanGenie (called from eight individuals with haplotype-resolved assemblies) and Sniffles (called from 25 HiFi read samples). (B) SV saturation for 25 HiFi read samples. Markers indicate the mean value of unique SVs over 10 random shuffles of sample order, and error bars represent the standard deviation. The dotted line is a fitted curve of the form f(x) = ax−b + c, predicting saturation at approximately 175,000 SVs. (C) SV overlap for different allele frequency (based on the 25 samples) bins. (D) Small variant accuracy of HiFi-based and short-read-based calls, taking the short-read data as truth, stratified by autosomes and sex chromosomes for SNPs and indels. Large markers indicate the mean over the 25 samples. (E) Small variant intersections between HiFi-based and short-read-based calls in genomic regions identified as centromeric satellites, low mappability, tandem repeats, repetitive, and “normal” (all other regions). A large proportion of variants called in the challenging regions were unique to HiFi-based alignment and calling.











