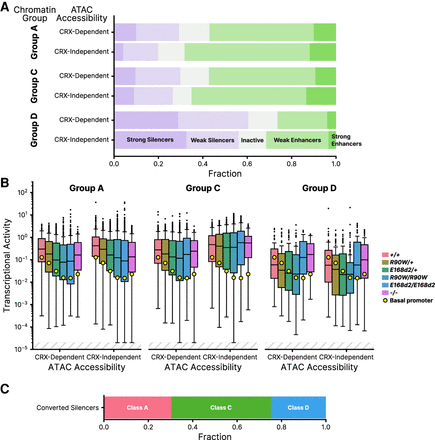
CRE activities analyzed by chromatin annotations at sites of genomic origin. (A) Distribution of CRE activity classes in +/+ retina separated into groups based on chromatin annotations of the site of genomic origin and CRX-dependent ATAC accessibility (Ruzycki et al. 2018). (Group A) H3K4me3+ H3K27ac+, (Group C) H3K4me3− H3K27ac+, (Group D) H3K4me3− H3K27ac−. (B) Transcriptional activity of all CREs, separated by chromatin and ATAC group, across all genotypes. (C) Chromatin classification breakdown of E168d2/d2 converted silencers (elements classified as weak or strong silencers in +/+ that become weak or strong enhancers in E168d2/d2). In panel B, transcriptional activity was adjusted for visualization purposes using a fixed pseudocount of 2 × 10−5; the out-of-bound region is indicated by the light gray stripes.











