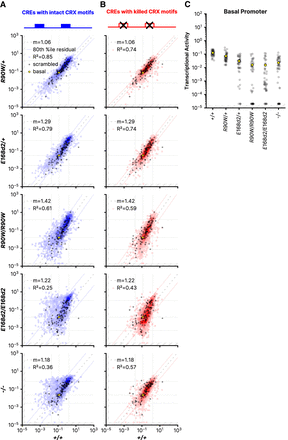
CRE activity in retinas with different Crx genotypes. (A,B) Correlations of the transcriptional activity of each CRE with intact CRX motifs (A) or CRE with killed CRX motifs (B) in the indicated genotypes. Superimposed black cross symbols indicate the transcriptional activity of the 100 scrambled control sequences, and a yellow circle shows the activity of the basal Rho promoter alone. Using the scrambled control sequences to establish an empirical null distribution of CRE activity, we performed robust regression using a trimmed mean M-estimator (fit plotted as a dashed line) for each genotype comparison. The outer dotted lines indicate the 80th percentile of the residuals of the library CREs relative to the regression of the scrambled sequences. The outer vertical and horizontal lines indicate the thresholds for the “strong silencer” and “strong enhancer” classes, and the inner horizontal and vertical lines correspond to the basal Rho promoter alone. (C) Activity of the Rho basal promoter construct in each of the indicated genotypes. Each black dot indicates a different unique sequence barcode; the mean across barcodes is shown as a yellow dot. In all panels, transcriptional activity was adjusted for visualization purposes using a fixed pseudocount of 2 × 10−5; the out-of-bound region is indicated by the light gray stripes.











