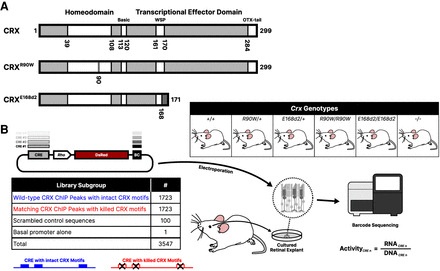
Study overview. (A) Schematics of wild-type CRX protein and the p.R90W and p.E168d2 pathogenic variants, containing a point DNA-binding domain mutation or resulting in truncation of the transcriptional effector domain, respectively. (B) Outline of the experimental procedure for constructing and testing the CRE libraries. A library of CRE sequences is cloned upstream of a rod photoreceptor-specific Rhodopsin (“Rho”) minimal promoter driving expression of the DsRed fluorescent protein, with each CRE marked by a unique sequence barcode (BC) in the 3′ UTR. Each library was electroporated into retinal explants with the six indicated Crx genotypes. RNA was collected from the retinas, and transcript counts for each element were measured and normalized to abundance in the input DNA library to calculate transcriptional activity.











