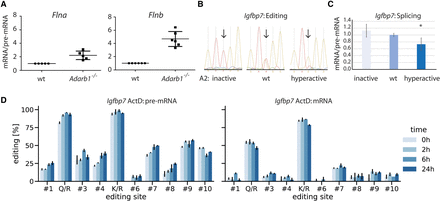
Editing sites close to 5′ splice sites and also distant from splice sites interfere with mRNA splicing. (A) RNA was isolated from wild-type and Adarb1−/− mice. Subsequently, pre-mRNA splicing efficiency was determined using qPCR for the editing targets Flna and Flnb. n = 5. (B,C) An Igfbp7 reporter construct was cotransfected into HEK293 cells either in combination with a construct expressing inactive ADARB1, a wild-type ADARB1, or a hyperactive version of ADARB1. Following isolation of RNA, editing levels (K/R site) were determined (B) and splicing efficiencies of the transcript in the presence of either ADARB1 was calculated (C). (D) An Igfbp7 construct was cotransfected with wild-type ADARB1 into HEK293 cells. Subsequently, transcription was blocked with actinomycin D (ActD). Using Sanger sequencing, editing levels for pre-mRNA and mRNA were determined at time points 0 h, 2 h, 6 h, and 24 h after ActD treatment (n = 3, error bars = standard deviation).











