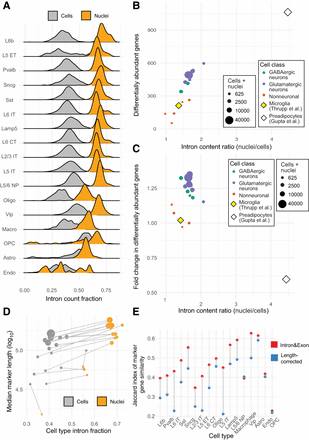
Variation in pre-mRNA enrichment moderates assay similarity with and without gene length normalization. (A) Intron content distribution (total Intron counts divided by total Intron&Exon counts per cell or nucleus) for each cell type in the mouse cortex. Mean intron content in cells does not increase with mean intron content in nuclei (Spearman's ρ = 0.19, P = 0.46). Nonneuronal cells are abbreviated: (astro) astrocytes, (endo) endothelial cells, (oligo) oligodendrocytes, (OPC) oligodendrocyte precursor cells, and (macro) macrophages. The remaining labels specify types of neuronal cells. (B) Differential expression testing of cells versus nuclei for each cell type. The number of differentially abundant genes (FC > 2) increases with the ratio of mean intron content in the cell type (Spearman's ρ = 0.89, P = 9.9 × 10−7). Cells are colored by cell type class. (C) Fold change in the number of differentially abundant genes for cells versus nuclei after applying the Gupta et al. length-correction procedure. The white diamond shows the result reported by Gupta et al. (2022) for white preadipocytes. Values greater than one indicate an adverse performance of the length-correction method. Linear modeling (of cortex cells only) identifies cell class and intron content ratio as significant predictors of method performance (R2 = 0.78, P = 0.0001). (D) Marker genes discovered from nuclei tend to be longer than those from cells of the same type, except for in very rare cell types. (E) Marker gene similarity (Jaccard index) for the top 50 markers (by fold change) is variable but not in relation to the number of cells or the intron content ratio. Columns are ordered by total (cells + nuclei).











