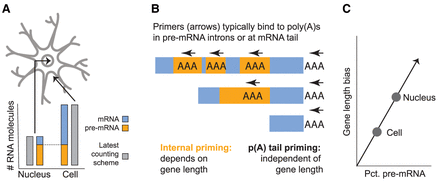
Conceptual depictions of key technical aspects of sc/snucRNA-seq. (A) Nuclei contain less total RNA but are enriched for pre-mRNA compared with cells. All pre-mRNAs in the nucleus are also in the cell, but most mRNAs are outside the nucleus. (B) Pre-mRNA tends to generate intronic reads via internal priming, which is biased in a gene length–associated matter. mRNA generates exonic reads from priming at the poly(A) tail, irrespective of length. Gray arrows indicate the position and direction of reverse transcription initiation by poly(dT) primers. (C) Gene length bias, or the degree to which long genes are overestimated by the assay, is a function of pre-mRNA content. Nuclei contain relatively more pre-mRNA than equivalent cells.











