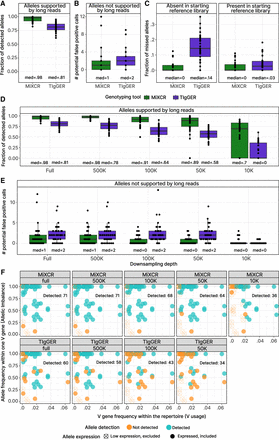
Detection of allelic variants of V genes by inference tools. (A) Fraction of allele calls supported by long-read-based genotyping. (B) Number of allele calls not supported by long-read-based genotyping. (C) Fraction of alleles, missed by MiXCR or TIgGER, by the presence in the initial reference library. (D,E) Sensitivity and specificity testing by downsampling each sample in the benchmarking data set by 500,000, 100,000, 50,000, or 10,000 reads. (D) Fraction of identified allele calls supported by long-read-based genotyping. (E) Number of identified allele calls not supported by long-read-based genotyping. (F) Detection of the allele variants of V genes depending on V usage and allelic imbalance. Each dot represents a V gene allele present in the donor's genotype confirmed by long-read sequencing. The upper row represents detection by the developed algorithm; the lower, allele detection by the comparison tool TIgGER. Columns represent different depths of downsampling by number of aligned reads, from right to left: full set of reads, 500,000, 100,000, 50,000, and 10,000. V gene, and allele frequencies for each facet were calculated using the full set of reads and allele-resolved V and J gene reference library. Alleles excluded due to low expression (<10 clonotypes), are represented as empty crossed points. N = 33 for A–E.











