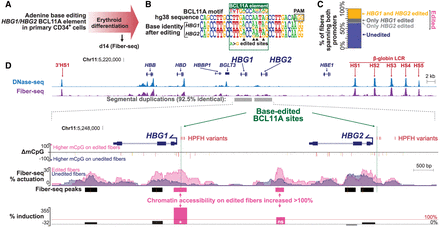
Therapeutic base editing of a BCL11A element within the HBG1/HBG2 promoters. (A) Schematic of the experimental paradigm. (B) Sequence logo showing base editing within the BCL11A binding site in the HBG1 and HBG2 promoters. Letter height corresponds to the relative base frequency. (C) Percent of fibers with edited BCL11A sites in both HBG1 and HBG2 (yellow), HBG1 only (dark gray), HBG2 only (light gray), and neither (blue). The top three categories are grouped as “edited” reads in D. (D) UCSC browser tracks of ABE-edited CD34+-derived erythroid cells. (Top) Comparison of ENCODE DNase-seq of treated multipotent progenitor cells (blue) and chromatin accessibility (FIRE) of CD34+ derived erythroids (purple) across all fibers mapping within the targeted region. (Bottom) Zoom-in with comparison of edited and unedited fibers across HBG1 and HBG2. The difference in CpG methylation is compared (unedited minus edited, yellow—P < 0.01, orange—P < 0.001, red—P < 0.0001, Fisher's exact test), as well as percent actuation of unedited fibers (blue), and HBG1 and/or HBG2 edited fibers (pink). Peak calls for edited reads and percent induction at each peak ([edited percent actuated − unedited percent actuated]/unedited percent actuated) are displayed at the bottom. Pink peaks and bars represent >100% induction. (*) indicates statistical significance (Fisher's exact test with Benjamini–Hochberg corrected FDR < 5%) (Supplemental Fig. S9).











