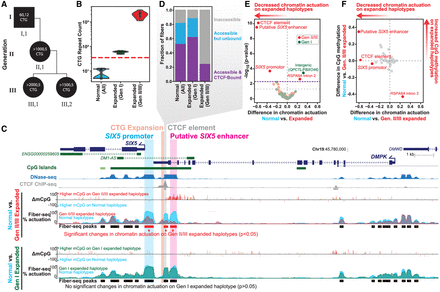
Haplotype-resolved chromatin accessibility and CpG methylation at the DMPK 3′-UTR CTG expansion in DM1 fibroblasts. (A) Family pedigree representing fibroblast donors. Labels inside each shape represent the CTG copy number within the pathogenic and normal allele, respectively. (B) CTG count across sequencing reads that fully span the CTG repeats, grouped by haplotypes. The red dashed line at 35 represents the threshold above which CTG expansions are unstable. (C) Browser tracks comparing the chromatin architecture of the normal haplotypes to the expanded haplotypes from generations II/III and generation I. The difference in CpG methylation is compared (yellow—P < 0.01, orange—P < 0.001, red—P < 0.0001, Fisher's exact test), as well as percent actuation of the normal (light blue), generations II/III expanded (red), and generation I expanded (green) haplotypes. Fiber-seq peaks from the normal haplotypes are shown below and are colored to represent a statistically significant decrease in chromatin accessibility on the generation II/III haplotypes (red) compared to normal. There were no significant changes between the normal and generation I (green) expanded haplotype. ENCODE DNase-seq and CTCF ChIP-seq are above in blue and gray, respectively. (D) CTCF footprinting at the CTG-adjacent CTCF-binding site. Footprints were classified as accessible if they were fully overlapped by a methylation-sensitive patch, and accessible footprints were classified as CTCF bound if they did not contain any m6A (P = 0.010, Fisher's exact test comparing CTCF bound to unbound [inaccessible plus accessible but unbound]). (E) Volcano plot of chromatin actuation difference at each accessible peak from the normal haplotype peak set within the targeted locus, compared between normal and expanded fibers from generation I (green) and generations II/III (red). The two peaks with increased accessibility in expanded fibers (upper right quadrant) are likely explained by SNPs local to each region (Supplemental Note). P-values were calculated by Fisher's exact test. The gray dashed horizontal line indicates the nominal significance threshold (P < 0.05), and the purple dashed line indicates the Benjamini–Hochberg FDR-corrected significance threshold. (F) Plot of CpG methylation difference versus chromatin actuation difference at the peaks described in E. Significant and nonsignificant points from E are shown in red and gray, respectively.











