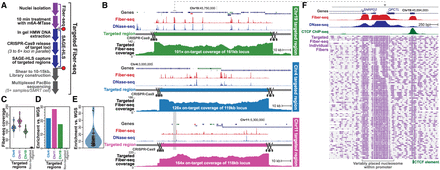
Targeted Fiber-seq methodology. (A) Schematic of targeted Fiber-seq protocol. (B) Fiber-seq percent actuation (red), ENCODE DNase-seq (blue), and per-base targeted Fiber-seq coverage are shown across targeted loci. (C) Violin plot of per-base coverage over the targeted regions, compared to a nontargeted region (Chr14:20283833–20402650, hg38). (D) Relative enrichment of targeted loci relative to WGS (see Methods). (E) Relative enrichment across all samples and targets. (F) Zoom-in of an illustrative locus showing m6A events (purple ticks) across individual fibers.











