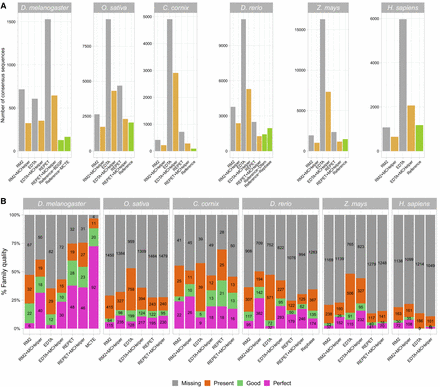
Comparison of MCHelper libraries with raw and reference libraries in the six genomes analyzed. (A) Number of consensus sequences contained in the raw library, the MCHelper library, and the reference libraries of the six species analyzed. Reference libraries used are: BDGP (Kaminker et al. 2002) and MCTE (Rech et al. 2022) for D. melanogaster, standard library (Ou et al. 2019) for O. sativa, MClibrary (Weissensteiner et al. 2020) for C. cornix, Dfam (Storer et al. 2021a) and Repbase (Jurka et al. 2005) for D. rerio, MTEC curated by Ou et al. (https://github.com/oushujun/MTEC) for Z. mays, and Dfam for H. sapiens. (B) Number of consensus sequences classified as perfect, good, present, and missing families following the methodology proposed by Flynn et al. (2020). Note that for D. melanogaster and D. rerio, the MCTE and the Repbase libraries are also compared to the BDGP and Dfam libraries, respectively.











