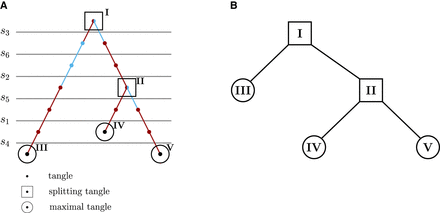
Tangles trees for the minimal example. (A) Tangles search tree. The constructed tangles search tree for the minimal example for agreement parameter a = 2. Cuts, sorted by cost, are evaluated sequentially, starting with the lowest-cost cut (s3). A cut is added to the tangles tree if it can be consistently oriented with respect to the previous cuts and their orientations, forming a tangle. The color indicates whether the orientation of the last added cut points to all individuals with at least one derived allele (light blue) or in the opposite direction (red). When a cut can be oriented in both directions (s3 and s5), it creates a split at that node, resulting in a splitting tangle (squares). The cut s4 cannot be added to every branch of the tangle tree. If no cut can be consistently added at any node or all cuts have been added to the tangles tree, the tangles tree is considered complete. The leaves represent the maximal tangles (circles). (B) Condensed tangles tree. Condensed tangles tree for agreement parameter a = 2, that is the tangles search tree from A reduced to splitting and maximal tangles.











