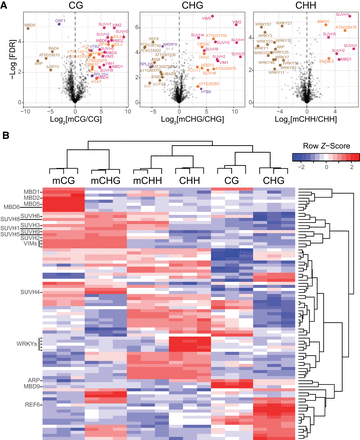
Identification of mC reader proteins in Arabidopsis from affinity pull-downs. (A) Volcano plots of the proteins statistically enriched for binding to methylated probes (known mC binders are pink, previously unknown mC binders are orange) or unmethylated probes (brown) in each context (CG, CHG, CHH). Statistically enriched proteins, often identified in mass spectrometry experiments and thus listed as contaminants in Van Leene et al. (2015), are colored purple. (B) Hierarchical clustering of the proteins (rows) statistically significantly enriched in at least one set of probes (columns, each representing a replicate). The names of proteins of interest were highlighted, the rest of the 83 proteins identified can be found in Supplemental Table S2.











