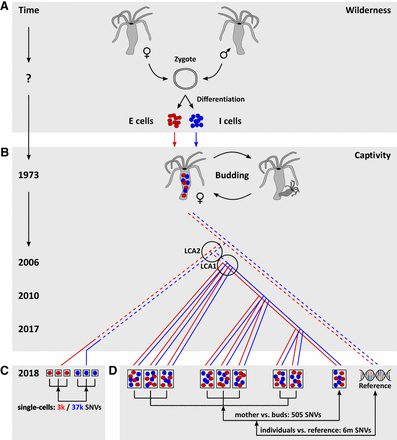
Biological and historical background, experimental setup, and pedigree relationships of analyzed genomic data sets of H. magnipapillata strain 105. (A) The last sexual reproduction, representing the strain's genetic origin, occurred in the wild. Shortly after fertilization, a Hydra zygote differentiates into noninterchangeable endodermal and ectodermal epithelial (E cells, red) and interstitial (I cells, blue) stem cell lineages. (B) The H. magnipapillata strain 105 was derived from a single female polyp that was caught in the wild in 1973 (Chapman et al. 2010; Schaible et al. 2015). The strain has since reproduced exclusively asexually by budding and has been propagated in laboratories worldwide. For generating a reference genome sequence, the strain was recloned from a single polyp in the laboratory of Dr. Hans Bode at UC Irvine in 2004 (Chapman et al. 2010). We obtained the strain from UC Irvine in 2006 and established a pedigree consisting of its mother and eight daughters (buds) generated at three time points: 2006, 2010, and 2017. (C) In 2018, we used single-cell whole-genome sequencing of three E cells and three I cells to identify single-nucleotide variants (SNVs) that had accumulated in each of these cells since their common origin (zygote). (D) In addition, we performed whole-animal sequencing of nine Hydra individuals, comprising the pedigree mentioned above. We identified SNVs that arose in each daughter in comparison to the mother. To further identify heterozygous genome positions of the strain, we also compared the data of all nine polyps individually with a reference genome sequence determined in another laboratory from more distantly related individuals (Chapman et al. 2010). The total numbers of uniquely identified SNV positions are displayed: in C for the comparisons of each E cell versus all I cells combined (red) and each I cell versus E cells combined (blue); in D for the comparisons of buds versus mother and individuals versus reference. In our experimental setting, the last common ancestor (LCA) 1 of the nine-individual pedigree was the mother at its stage of somatic evolution in 2006. The LCA2 of our nine-individual pedigree and the individual used for single-cell sequencing was budded also in 2006. The LCA of the individuals sequenced by us and those from which the reference genome was derived can be dated back only to a time window of 1974–2004. Pedigree lines: continuous—life history of a Hydra individual; dashed—descant relation including multiple Hydra individuals derived from each other by budding.











