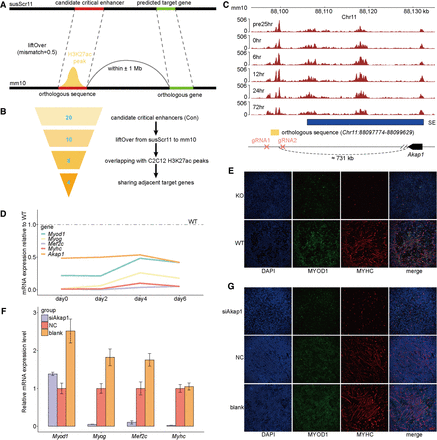
Conservation analysis and functional assay of candidate critical enhancers within Con SEs. (A) Schematic diagram illustrating the mapping of candidate critical enhancers to functional orthologous sequences in the mouse genome. (B) Flowchart illustrating the conservative filtering steps identifying functional orthologous sequences of candidate critical enhancers within Con SEs. The Arabic numerals indicate the number of candidate critical enhancers retained in each filtering step. (C) Genome browser snapshot displaying the mouse orthologous sequence of the candidate critical enhancer Chr12:34230720–34231974, extended 500 bp upstream and downstream. The tracks represent H3K27ac ChIP-seq signals across six stages during C2C12 myogenesis. The 24 h before induction of differentiation are denoted by “pre24hr,” and so forth. The gRNAs targeting the sequence and the predicted target gene were annotated. (D) qPCR analysis of the relative expression of four myogenic markers (Myod1, Myog, Mef2c, and Myhc) and one predicted target gene (Akap1) in wild-type (WT) and knockout (KO) C2C12 cells at four myogenic differentiation time points. KO cells were generated by deleting the orthologous sequence using CRISPR–Cas9. All qPCR data are presented as means ± SD of triplicate experiments. (E) Immunofluorescence staining of WT and KO C2C12 cells after 6 days of differentiation induction. Differentiated myotubes were labeled by immunostaining for MYOD1 (green) and MYHC (red), with nuclei stained by DAPI (blue). (F) qPCR analysis of the relative expression of myogenic markers in C2C12 cells with Akap1 knockdown on day 7 of myogenic differentiation. (G) Immunofluorescence staining for MYOD1 and MYHC was used to assess the effect of Akap1 knockdown on C2C12 differentiation.











