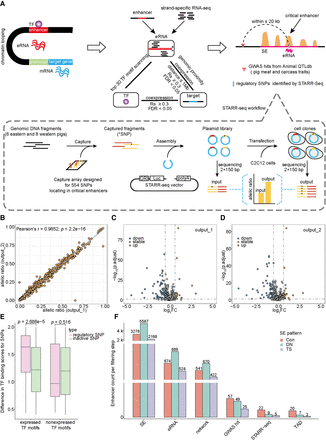
Screening strategy for candidate critical enhancers. (A) Candidate critical enhancers were defined as SE units meeting three conditions: transcribing eRNAs, overlapping with GWAS hits related to pig meat and carcass traits (trait classes from Animal QTLdb), and harboring eRNA-located GWAS single-nucleotide polymorphisms (SNPs) that modulated enhancer activity in STARR-seq experiments. (B) Correlation of allelic ratios for SNPs between two STARR-seq biological replicates. (C,D) Identification of enhancer-modulating regulatory SNPs by the change in allelic ratios between the output library, including replicate1 (C) and replicate2 (D), and the input library. Statistical significance (false discovery rate [FDR] < 0.05) was evaluated using Fisher's exact test. (E) DNA-binding motif-breaking scores were assessed for regulatory SNPs and inactive SNPs on binding motifs of TFs expressed (TPM ≥ 3 at least at one stage) and nonexpressed (TPM < 3 at all stages) in PSM. (F) Number of enhancers retained during candidate critical enhancer screening process. (TAD) topologically associating domain.











