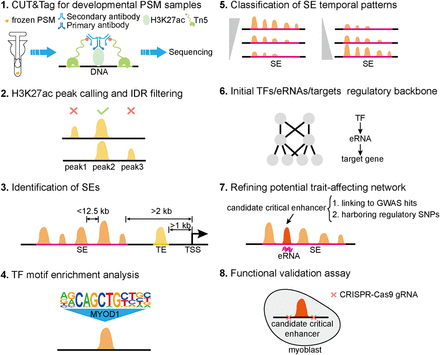
Workflow for revealing potential critical enhancers and trait-affecting gene regulatory networks during PSM development. (1) CUT&Tag libraries from frozen PSM samples across five developmental stages are produced and sequenced. (PSM) porcine skeletal muscle. (2) H3K27ac peaks are identified at each developmental stage and filtered for intragroup consistency. (IDR) irreproducibility discovery rate. (3) Identify SEs and typical enhancers (TEs) based on distal H3K27ac peaks. (ROSE) ranked order of super-enhancers, (TSSs) transcription start sites. (4) Analysis of enriched TF motifs on SEs. (5) Using hierarchical clustering to reveal temporal patterns of SEs. (6) Construct gene regulatory networks mediated by potential critical enhancers based on TF motif scanning and coexpression relationships among TFs, eRNAs, and target genes. (7) Refinement of trait-affecting regulatory network by identifying candidate critical enhancers within SEs, characterized by eRNA transcription and GWAS variants modulating enhancer activity. (8) Validate the role of candidate critical enhancers in myogenic differentiation using CRISPR–Cas9-mediated enhancer knockout in C2C12 cells.











