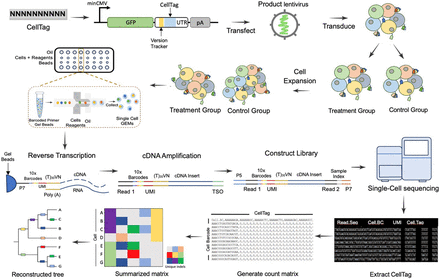
Standard workflow of single-cell lentivirus barcoding lineage tracing experiment. Lentivirus barcode labeling: cells of interest are labeled with DNA barcodes through lentivirus transfection. Cell expansion and splitting: After reaching the required cell numbers, cells are split for drug treatment or challenges, with one portion saved as a parental control. Single-cell RNA sequencing (10x Genomics): 10x Genomics is employed for scRNA-seq, where CellTag sequence can be captured from the 3′ UTR of fluorescence gene similar to other genes. This step involves Gel Bead Emulsions, reverse transcription, amplification, and sequencing. Computational analysis: DNA barcodes are used in computational analysis to connect cell fate after treatment or challenge to its origin based on clone calling and lineage tree inference. For example, the summarization matrix and the lineage tree show that cells A, B, and C have the same barcodes and they are inferred to have a closer relationship from lineage analysis, so do cells E, F, and G. Molecular mechanism investigation: Single-cell transcriptomics data are utilized to understand the molecular mechanisms underlying fate determination.











