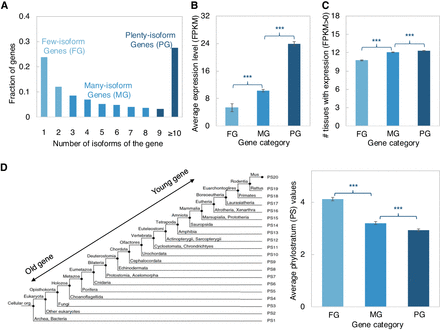
Characteristics of gene loci with different numbers of isoforms. (A) Gene categories are based on the number of isoforms of each gene. Three gene categories were classified with roughly the same number of genes in each group. (B) Comparison of the average gene expression level among the three gene categories. The average gene expression level was calculated based on the Illumina RNA-seq generated in this study. (C) Comparison of the number of tissues with expression among the three gene categories. The transcriptomic sequencing data from 13 tissues of the M. musculus C57BL/6 inbred line was retrieved from Lin et al. (2014). The detectable gene expression in each tissue was defined as FPKM > 0. (D, Left) Depiction of the phylostratum (PS) assignments for mouse protein-coding genes were retrieved from Neme and Tautz (2013). (Right) Comparison of the average PS values among the three gene categories. The smaller PS values indicate the older gene groups. Error bars show SEM values of average gene expression levels, number of tissues with expression, and average PS values, respectively. Statistical P-values in all panels were computed using two-sided Wilcoxon rank sum tests; (***) P-value < 0.001, (**) P-value < 0.01, (*) P-value < 0.05, and (ns) P-value ≥ 0.05.











