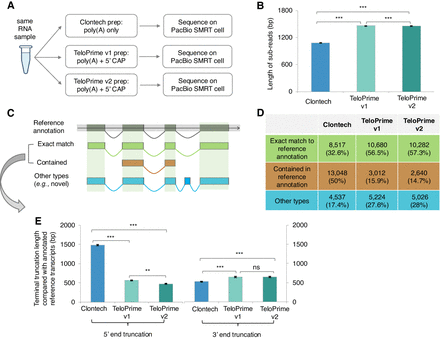
Performance evaluation of three types of full-length transcript enrichment protocols. (A) A simplified flow chart of this evaluation experiment setup. (B) The length distribution of subreads generated from three PacBio Iso-Seq libraries. (C) The major categories of Iso-Seq transcripts based on their alignment coordinates to the reference genome, and (D) for the distribution of these categories. (E) The terminal truncation length of Iso-Seq transcripts from both 5′ and 3′ ends, compared to the reference annotation in Ensembl v103. Only Iso-Seq transcripts with exact matches or wholly contained in reference annotation were taken for analysis. The error bars indicate the standard error of mean (SEM) of the length values. Statistical P-values were computed using two-sided Wilcoxon rank sum tests; (***) P-value < 0.001, (**) P-value < 0.01, (*) P-value < 0.05, and (ns) P-value ≥ 0.05.











