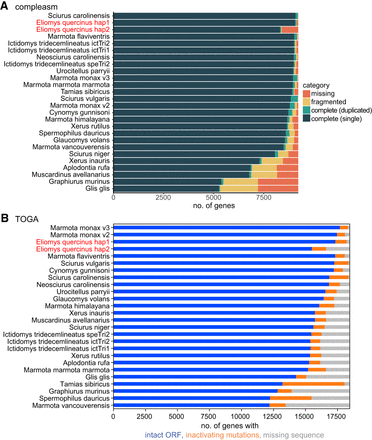
Comparison of gene completeness between our haplotype-resolved garden dormouse and other Sciuromorpha genomes. (A) Fractions of detected, fragmented, and missing BUSCO genes (mammalian odb10 data set, 9226 genes), as classified by compleasm v.0.2.2 (Huang and Li 2023) across Sciuromorpha assemblies. (B) TOGA classification of 18,430 ancestral placental mammal genes across Sciuromorpha assemblies. Assemblies are ranked by the number of completely detected (A) and by the number of intact (B) genes. For the garden dormouse, we show both haplotypes in red font and next to each other, but the X Chromosome is only contained in haplotype 1. Species and assembly accessions are listed in Supplemental Table S1.











