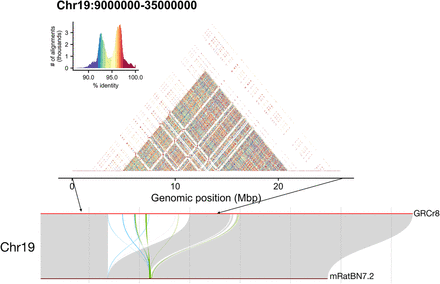
Alignment of Iso-Seq data from several BN tissues obtained from the MCW suggested that the newly incorporated region on Chr 19 shows aligned transcript data across its extent. This is in contrast to newly incorporated centromeric regions in which no aligned transcripts were observed. To investigate further, we examined whether the included region might be composed of expanded genomic sequences with high similarity, such as might occur as the result of repeated segmental duplication. Self-to-self similarity visualization with StainedGlass reveals a highly duplicated structure that lacked the simple expansion observed in centromeric regions. This newly incorporated region of Chr 19 contains 276 previously unmapped RefSeq genes and 12 additional proposed genes from Gnomon annotation analysis.











