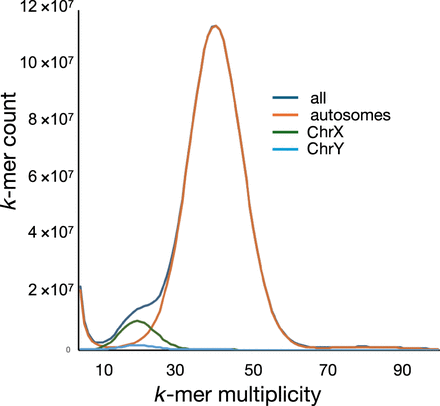
We extracted all 21-mers from the HiFi reads to evaluate the k-mer distribution in the assembly. k-mers were aligned to the autosomes, extracted, and plotted to demonstrate multiplicity of k-mers. As anticipated, autosomally mapped k-mers show a single peak with a multiplicity similar to the sequencing read depth, indicative of the absence of heterozygosity. Similar mapping and plotting was done for the sex chromosomes, which both show a multiplicity indicative of their haploid state in the male subject genome. Supplemental Figure S2 shows each plot (autosomes, Chr Y, and Chr X) independently as well as the combined plot of all k-mers, allowing closer examination.











