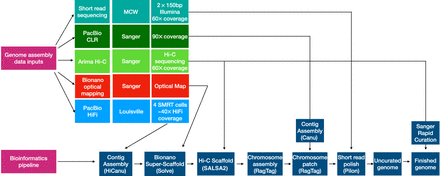
Genome assembly pipeline and data inputs. Illumina short-read sequencing was generated by the Medical College of Wisconsin (MCW) and used for assembly polishing. PacBio CLR reads generated by the Wellcome Sanger Institute and originally used for creating the preceding reference assembly mRatBN7.2 were assembled with Canu, and the resulting contigs were used for assembly gap filling. Scaffolding was performed using Bionano Genomics optical reads and Arima Genomics Hi-C data generated at Sanger for the mRatBN7.2 assembly. PacBio HiFi reads were generated on a Sequel IIe instrument at the University of Louisville. These were assembled with HiCanu, and the resulting contigs were integrated with the Bionano optical map to create a hybrid assembly using Bionano Solve software at Texas A&M University. The Hi-C data were used to further scaffold the Bionano hybrid assembly using SALSA2 software. Reference-assisted chromosome assembly used RagTag software, and the existing mRatBN7.2 assembly served as a template to order and assemble scaffolds, into which the PacBio CLR contigs were incorporated for gap filling using the RagTag patch module. After polishing, the assembly was curated using the Sanger Institute's rapid curation pipeline, which uses Hi-C contact mapping to identify and resolve misassemblies.











