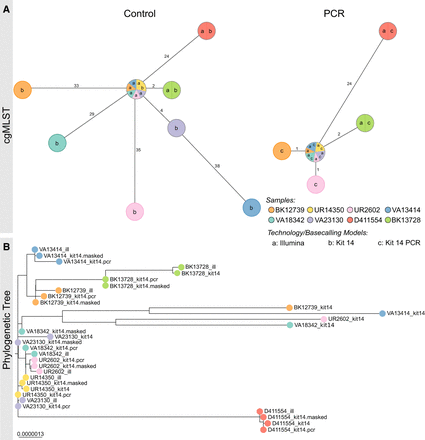
PCR-based sequencing or masking of ambiguous positions reduces allelic or phylogenetic distances. (A) Minimum spanning trees (pairwise ignore missing values) of each of eight K. pneumoniae outbreak samples based on 2358 genes to compare allelic differences between Illumina and Nanopore SQK-NBD114.24 genomes (Kit 14; left) and Illumina and Nanopore SQK-RPB114.24 genomes (PCR; right). Nodes (samples) are connected by lines depicting the distance by numbers of allelic differences. Loci are considered different whether one or more bases change between the samples. Loci without allelic differences are described as being the same. Samples with allelic differences of 15 or fewer are considered as part of the cluster. (B) Phylogenetic tree based on core genome SNP alignment between eight K. pneumoniae outbreak samples (colored nodes), prepared with Illumina (ill), Nanopore SQK-NBD114.24 (Kit 14), and SQK-RPB114.24 (Kit 14 PCR) compared with the masked Kit 14 assemblies (masked).











