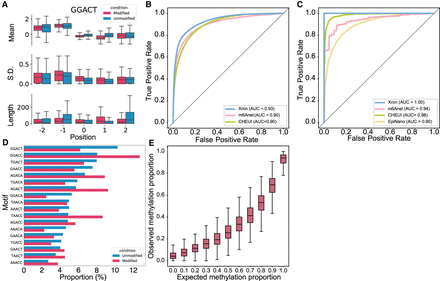
Evaluation of the m6A detection results obtained for synthesized IVT RNA reads and stoichiometry prediction. (A) Box plot comparing the distribution of the mean, standard deviation, and length for the signal segmented by NHMM with 5232 modified sites and 18,464 unmodified sites for the GGACT motif. Horizontal lines show the median, the box denotes the interquartile range, and the whiskers extend to 1.5 times the interquartile range. Points beyond this range are considered outliers and are removed from the plot. (B,C) ROC curves of Xron against m6Anet and CHEUI for read-level (B) and site-level (C) m6A modification predictions. (D) Bar plot showing the relative proportion of DRACH 5-mer motif for 84,919 modified and 179,717 unmodified positions. (E) Box plot showing the m6A ratio predicted by Xron with different proportions of IVT control and IVT m6A RNA mixing.











