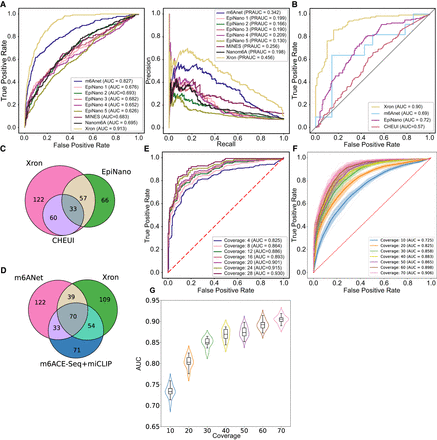
Comparison of Xron models across two different species. (A) ROC and PR curves of m6A prediction on human HEK293T cell line, produced by Xron and other models. (B) ROC curves produced by Xron and other models on yeast data. (C,D) Venn diagram showing the overlapping sites predicted by Xron and other methods on yeast (C) and HEK293T (D) data. (E) ROC curves produced by Xron for detecting m6A methylation in yeast data under different minimum sequence coverage thresholds. (F) ROC curves generated by Xron for detecting m6A methylation in down-sampled yeast data with different coverage. (G) Distribution of AUC score of Xron on down-sampled yeast data.











