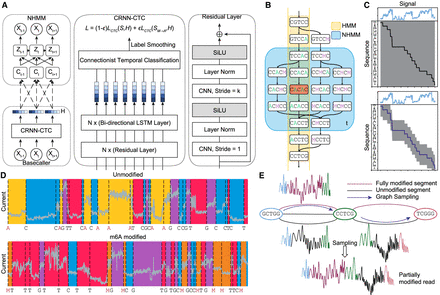
Schematics of Xron model and the data augmentation process through cross-linking and sampling. (A) Xron consists of two parts: a NHMM and a CRNN with a connectionist temporal classification (CTC) decoder. (B) Comparison between HMM and NHMM. The transition matrix of a HMM (yellow) encodes the whole Markov chain of k-mers, while the transition matrix of the NHMM (blue) at time t only encodes the Markov chain of the five nearby k-mers given the predicted k-mer (shown in red) at time t. The Markov chain is also expanded to include the k-mers with all combinations of the A and M (m6A) bases. We create partially methylated reads using data augmentation, first segmenting the signal and then cross-linking the reads and their corresponding signal in silico. To achieve this, we design a novel NHMM that can be trained to conduct signal segmentation in a semisupervised fashion on modified reads, even when lacking methylation labels. The NHMM is trained using the forward–backward algorithm with its transition matrix conditioned on a canonical basecalled sequence and its alignment, thus giving the maximum likelihood estimation of the model parameters regarding the methylation base. The Viterbi path of the NHMM gives the alignment between the current signal and sequence. Following the signal segmentation process performed with the NHMM, the NHMM was used to create a training data set with partially methylated reads and their true labels for methylation detection training by augmenting all-or-none modified reads. (C) The transition process of the NHMM is constrained by the neural network's output, leading to a smaller probability space and making it easier for the model to find the optimal alignment. (D) The NHMM is trained in a semisupervised manner on IVT data sets, including fully modified, unmodified, and partially modified reads. It provides accurate signal segmentation results for both unmodified and modified sequences. (E) In silico read cross-linking. The fully modified or unmodified reads are first broken into segments at the invariant k-mers to form a signal-k-mer graph, whose nodes are k-mers and whose edges are signal segments. Then, a partially methylated read is sampled from the k-mer signal graph.











