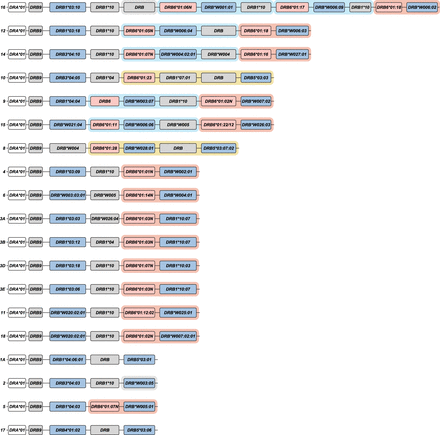
A schematic representation of the composition of distinct rhesus macaque DR region configurations. The different region configurations analyzed in this cohort are numbered according to a previous report (Doxiadis et al. 2013). Functional genes are indicated in blue boxes, whereas pseudogenes are in gray. These inactive genes are designated based on the functional allele with which they exhibit at least 95% sequence similarity, whereas inactive entities lacking similarity are denoted as “DRB.” Pseudogene DRB6 is an exception and is depicted with red boxes. The DRB9 and DRA loci (white boxes) represent a framework that is structurally consistent on every region configuration. The locus equivalent to HLA-DRB1 is often occupied by a homolog, but other genes are encountered as well. The shadings in four different colors, connecting multiple gene boxes, indicate the different cassettes of paralogous genes. The cassettes are distinguished by their gene content, which is indicated in Figure 6. These cassettes always start with a DRB6 copy, which propels the region diversification via two instable retroviral elements. Two other genes, a truncated one on region configuration 16 (DRB) and a functional gene on configuration 2 (DRB*W003:05), also comprise one of these LTRs and might form a rearranging cassette on their own (depicted with a gray shading).











