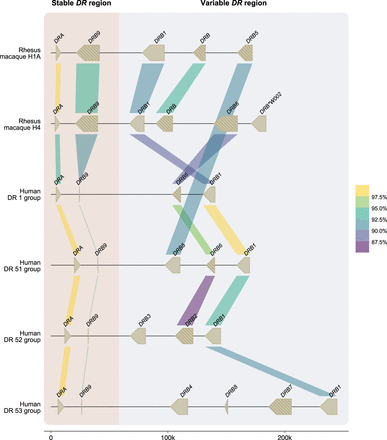
A schematic overview of the similarity between human and rhesus macaque DR genes. Two rhesus macaque DR region configurations (1A and 4) are depicted at scale, which are assembled using a combination of ONT and PacBio data (>60 × coverage). For comparison, four human DR region configurations, representing the DR1, DR51, DR52, and DR53 group haplotypes, were extracted from the NCBI database (GRCh38.p14: NC_00006, NT_167249.2; OK649233; NT_113891) (Houwaart et al. 2023). The DR genes and their orientations are represented by arrows. A striped pattern indicates pseudogenes. The color-coded bars connect homologous human and macaque genes and indicate their sequence similarity percentages that are determined using BLASTN (see Methods) (Altschul et al. 1990). The DRA and DRB9 loci represent a stable stretch in the DR region, whereas a more diverse gene content is encountered for the remaining haplotype. In the selected macaque and human region configurations, four DRB genes were determined to be orthologs, with similarities up to 90%–93%, whereas the DRA gene is even more conserved. The rhesus macaque MHC class II genes were originally named based on resemblance of their exon 2 sequences to HLA equivalents (de Groot et al. 2012). In the case that such an equivalent was absent in humans, a W (workshop) assignment was introduced, for instance, for Mamu-DRB*W002.











