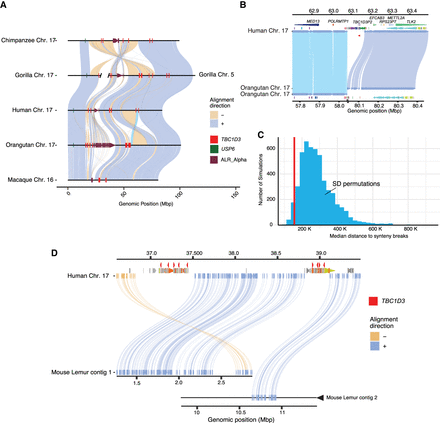
Large-scale chromosomal rearrangements and TBC1D3 duplications. (A) Synteny plots of orthologous Chromosome 17 in primates reveal syntenic blocks in direct (blue) and inverted (yellow) orientation. Alpha satellite sequence, TBC1D3 copies, and USP6—a hominoid fusion gene of TBC1D3—are illustrated in maroon, red, and green, respectively. TBC1D3 demarcates the boundaries of large-scale rearrangements on chromosome phylogenetic group XVII. (B) TBC1D3 duplication block (cluster of colored arrows) demarcates the boundary of a 12 Mbp inversion between the human and orangutan chromosomes. (C) Permutation test of segmental duplication proximity to synteny breaks. Five thousand permutation tests were performed, in which segmental duplication samples were taken, and median proximity to breaks in synteny was measured. True TBC1D3 mappings fall within the lowest 3% of the permutations (red line), suggesting a nonrandom association between TBC1D3 and breakpoints in synteny. (D) Synteny plot showing orthologous alignments between human TBC1D3 and mouse lemur flanking genomic sequence.











