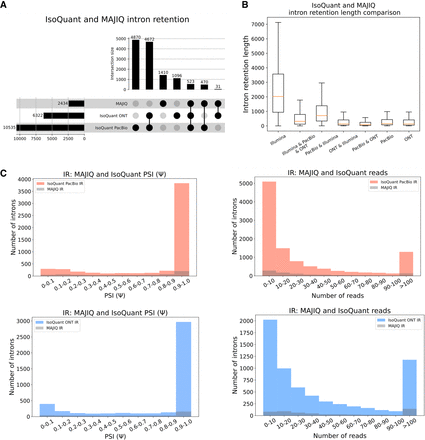
Comparison of IR events. (A) Upset plot showing overlap and total IR events reported by MAJIQ from short reads and IsoQuant using PacBio- or ONT-matched long reads (LRGASP data set). (B) Boxplots showing IR length distribution across seven categories in A. Each boxplot represents the IR length (y-axis) in each category (x-axis). The median is denoted by the yellow line; the upper and lower quartiles are denoted by the box; and the whiskers show points that lie within 1.5 IQRs of the lower and upper quartiles. The number of events in each category corresponds to those in A. (C) Introns reported by IsoQuant using PacBio (red) or ONT (blue) and how many of those were also identified by MAJIQ (gray) as a function of the PSI values (left) or the number of reads covering the intron (right). For PSI, only IR events with at least 10 reads were considered, and if an intron appears multiple times, the lowest PSI value is chosen for it.











