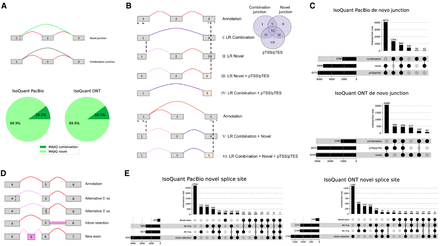
Analysis of de novo elements. (A) Short-read-only de novo splice junctions reported by MAJIQ (green junctions between exon 1 and 3 in the splicegraphs) can be classified as those involving novel splice sites (light green) or a novel combination of known splice sites (dark green). Red junctions correspond to annotated ones. The pie chart shows that compared with long reads processed with IsoQuant, ∼90% of MAJIQ de novo splice junctions involve novel splice sites. (B) Representative cartoon examples for six different categories of long-read de novo transcript variations. Novel combination junctions (dark purple), junctions involving novel splice sites (light purple), and junctions supported by annotation (red) are the same as in A. Putative starts or ends (pTSSs/pTESs; light yellow), or partial exons, represent cases when the transcript start sites or transcript end sites do not match those in the annotation, which happens in the first or last exon of the transcript. We note that cases involving only pTSSs/pTESs (class VII in the Venn diagram) are not included in downstream analysis as those are not handled by MAJIQ or similar short-read-based splicing algorithms, so they cannot be directly compared. (C) Breakdown of all cases involving de novo junctions reported by IsoQuant using either PacBio (top) or ONT (bottom) long reads. Notably, almost all of those cases also include pTSSs/pTESs. (D) Representative cartoon examples for the types of novel splice variations (pink) that a novel splice variant in long reads can introduce compared with the annotation (top graph, red junctions). (E) Breakdown of long-read novel splice junctions (light purple in B) into the four different categories shown in D when using IsoQuant to analyze PacBio (left) and ONT (right) matched reads.











