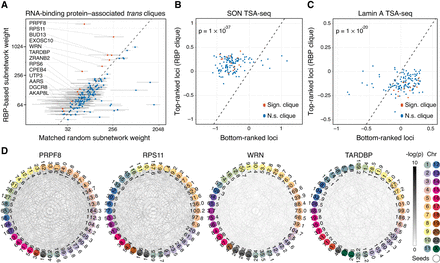
Trans-C identifies RNA-binding protein (RBP)–associated trans cliques proximal to nuclear speckles. (A) A subset of trans-C-identified subnetworks in lymphoblastoid cells built from loci characterized by strong binding of RBPs have denser contacts than the corresponding matched random null model. We plot the weight of a RBP-based subnetwork and the average weight of 1000 matched random seed subnetworks (error bars correspond to the SD) for 139 RBPs. In red and listed by name are those with significant P-values after FDR correction. (B,C) RBP-associated cliques identified by trans-C are significantly closer to nuclear speckles (stronger SON TSA-seq signal) and significantly further away from the nuclear lamina (weaker Lamin A TSA-seq signal) than matched control sets at the opposite end of the trans-C rankings. (D) Visualization of selected significant RBP-associated trans cliques in lymphoblastoid cells, plotted as described for Figure 2D.











