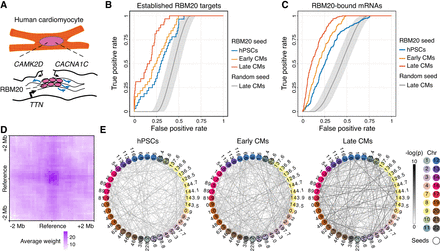
Trans-C identifies the RBM20 splicing factory in human cardiomyocytes (CMs). (A) Schematic of the RBM20 splicing factory, a muscle-specific interchromosomal structure organized by the TTN pre-mRNA. This pre-mRNA binds to more than 100 copies of RBM20 and nucleates foci that engage with other RBM20 targets to promote their alternative splicing (blue arrows). (B) Performance evaluation of trans-C in uncovering the RBM20 splicing factory in early (day 15) versus late (day 80) CMs differentiated from human pluripotent stem cells (hPSCs; also analyzed as “day 0” baseline control). Results for late CMs are statistically significant (P = 5 × 10−122) versus a matched random seed null model (average and 95% confidence interval from 1000 runs). Seed loci and ROC curves are based on a list of established RBM20 targets (Bertero et al. 2019). (C) Similar to B, but seed loci and ROC curves are based on loci directly bound by RBM20 as determined by eCLIP; P = 4 × 10−120. (D) Aggregated heatmap of trans contacts between the top 60 loci selected by trans-C in late CMs starting from eCLIP data. Each square in the grid represents an average 100 kb bin in a Hi-C matrix of 41 × 41 bins centered at each interacting pair of loci extracted from the Hi-C data (reference). The denser region in the middle reveals the specific nature of the trans interactions at the RBM20 splicing factory. (E) Visualization of the RBM20-associated trans clique identified by trans-C in late CMs starting from eCLIP data, showcasing the increased significance of loci interactions during hPSC differentiation and CM maturation, plotted as described for Figure 2D.











