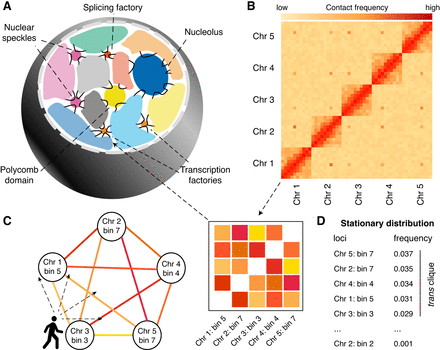
The trans-C algorithm. (A) Schematic of typical interchromosomal genome organization in mammals. Interchromosomal (trans) interactions mainly involve genomic domains that extrude from chromosome territories and engage with a variety of membrane-less structures involved in gene regulation. (B) A Hi-C matrix captures the contact frequency of loci in a genome-wide fashion. Besides intrachromosomal (cis) contacts, specific loci can exhibit strong interchromosomal (trans) contacts among themselves. (C) Trans-C employs a random walk algorithm that traverses the Hi-C contact graph choosing to move to a node (bin) probabilistically based on the strength of the edge (interaction). (D) The output is a list of loci ranked by how frequently they are visited during the random walk: More frequently visited loci interact more strongly in trans as a clique.











