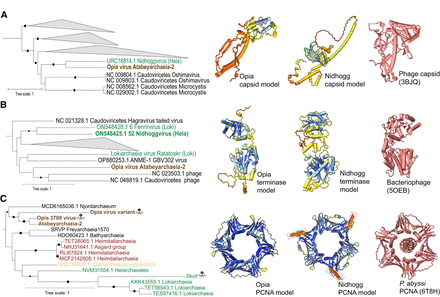
Evolutionary relationships and structural conservation of Opia virus hallmark proteins. (A–C) Maximum-likelihood phylogenetic trees of capsid (A), terminase large subunit (B), and proliferating cell nuclear antigen (PCNA; C). Trees are midpoint-rooted; circles indicate bootstrap support >70%. Insets show structural models: Opia virus (infecting Atabeyarchaeia, this study) and Nidhogg virus (infecting Helarchaeales) colored by AlphaFold3 confidence (blue to red, high to low), with best FoldSeek match in coral. Reference structures: bacteriophage capsid (PDB: 3BJQ) for A, large terminase from thermophilic bacteriophage D6E (PDB: 5OEB) for B, and P. abyssi PCNA (PDB: 6T8H) for C. Scale bars represent one substitution per site. Opia virus variants cluster with other Asgardarchaeota MGEs, highlighting their evolutionary relationships within this archaeal phylum.











