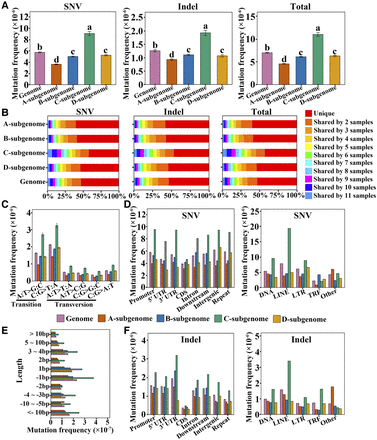
DNA mutations among the 12 “Beni hoppe” individuals. (A) SNV, indel, and total mutation frequencies (number of mutations per site) of the 12 “Beni hoppe” individuals relative to FaBen in the whole genome or each subgenome. Data are the mean ± SEM. Different letters indicate significant differences at P < 0.05 using Tukey's test. (B) Percentage of mutations shared by 12 samples in the whole genome or each subgenome. (C) Mutation frequencies (number of mutations per site) of different transitions or transversions in 12 samples. (D) Mutation frequencies (number of mutations/total length of each region type) of SNVs in different genomic regions of protein-coding genes or transposon families. (E) Mutation frequencies (number of mutations per site) of indels with different lengths. (F) Mutation frequencies (number of mutations/the total length of each region type) of indels in different genomic regions of protein-coding genes or transposon families. (Promoter) 2 kb upstream of transcription start site (TSS), (UTR) untranslated region, (Downstream) 2 kb downstream from transcription termination site (TTS), (Repeat) transposable element or tandem repeat sequence, (TRF) tandem repeat sequence; Other, unclassified transposon.











